| dyads_test_vs_ctrl_m2_shift1 (dyads_test_vs_ctrl_m2) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; dyads_test_vs_ctrl_m2; m=0 (reference); ncol1=11; shift=1; ncol=13; -wwaGGsCCAww-
; Alignment reference
a 0 563 666 1378 136 95 163 68 90 1861 744 694 0
c 0 404 312 144 35 36 1314 1911 1861 57 434 420 0
g 0 412 450 170 1839 1867 520 25 35 53 320 382 0
t 0 684 635 371 53 65 66 59 77 92 565 567 0
|
| TCP3_M1645_1.02_CISBP_shift2 (TCP3:M1645_1.02:CISBP) |
 |
0.791 |
0.719 |
3.980 |
0.916 |
0.929 |
1 |
1 |
19 |
1 |
1 |
4.600 |
1 |
; dyads_test_vs_ctrl_m2 versus TCP3_M1645_1.02_CISBP (TCP3:M1645_1.02:CISBP); m=1/27; ncol2=10; w=10; offset=1; strand=D; shift=2; score= 4.6; --kgGGaCCAch-
; cor=0.791; Ncor=0.719; logoDP=3.980; NsEucl=0.916; NSW=0.929; rcor=1; rNcor=1; rlogoDP=19; rNsEucl=1; rNSW=1; rank_mean=4.600; match_rank=1
a 0 0 17 17 3 3 62 2 3 80 8 29 0
c 0 0 24 5 2 12 24 96 97 7 67 29 0
g 0 0 31 58 91 80 3 2 0 11 11 16 0
t 0 0 28 20 4 5 11 0 0 2 14 26 0
|
| LOC_Os02g42380_M1657_1.02_CISBP_shift1 (LOC_Os02g42380:M1657_1.02:CISBP) |
 |
0.730 |
0.664 |
3.849 |
0.904 |
0.908 |
8 |
2 |
20 |
4 |
4 |
7.600 |
3 |
; dyads_test_vs_ctrl_m2 versus LOC_Os02g42380_M1657_1.02_CISBP (LOC_Os02g42380:M1657_1.02:CISBP); m=3/27; ncol2=10; w=10; offset=0; strand=D; shift=1; score= 7.6; -kkGGgcCCmm--
; cor=0.730; Ncor=0.664; logoDP=3.849; NsEucl=0.904; NSW=0.908; rcor=8; rNcor=2; rlogoDP=20; rNsEucl=4; rNSW=4; rank_mean=7.600; match_rank=3
a 0 18 7 0 0.0000 13 11 0.0000 7 45 28 0 0
c 0 13 15 0 0.0000 10 66 1.0000 93 33 41 0 0
g 0 41 33 93 1.0000 66 10 0.0000 0 15 13 0 0
t 0 28 45 7 0.0000 11 13 0.0000 0 7 18 0 0
|
| OJ1581_MA1031.1_JASPAR_shift1 (OJ1581:MA1031.1:JASPAR) |
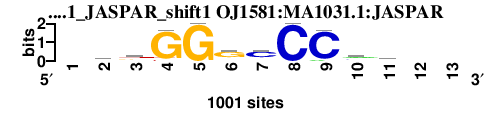 |
0.729 |
0.663 |
3.844 |
0.904 |
0.907 |
9 |
3 |
21 |
5 |
5 |
8.600 |
4 |
; dyads_test_vs_ctrl_m2 versus OJ1581_MA1031.1_JASPAR (OJ1581:MA1031.1:JASPAR); m=4/27; ncol2=10; w=10; offset=0; strand=D; shift=1; score= 8.6; -kkGGgcCCmm--
; cor=0.729; Ncor=0.663; logoDP=3.844; NsEucl=0.904; NSW=0.907; rcor=9; rNcor=3; rlogoDP=21; rNsEucl=5; rNSW=5; rank_mean=8.600; match_rank=4
a 0 175 74 0 0 127 112 0 71 445 279 0 0
c 0 131 146 0 0 101 661 1000 929 335 414 0 0
g 0 414 335 929 1000 661 101 0 0 146 131 0 0
t 0 279 445 71 0 112 127 0 0 74 175 0 0
|
| TCP23_TCP23_ArabidopsisPBM_shift2 (TCP23:TCP23:ArabidopsisPBM) |
 |
0.721 |
0.655 |
5.349 |
0.880 |
0.855 |
14 |
4 |
9 |
16 |
21 |
12.800 |
9 |
; dyads_test_vs_ctrl_m2 versus TCP23_TCP23_ArabidopsisPBM (TCP23:TCP23:ArabidopsisPBM); m=9/27; ncol2=10; w=10; offset=1; strand=D; shift=2; score= 12.8; --GGGGCCCACA-
; cor=0.721; Ncor=0.655; logoDP=5.349; NsEucl=0.880; NSW=0.855; rcor=14; rNcor=4; rlogoDP=9; rNsEucl=16; rNSW=21; rank_mean=12.800; match_rank=9
a 0 0 2 0 1 3 1 0 1 96 1 74 0
c 0 0 5 0 1 9 97 98 98 1 96 14 0
g 0 0 77 97 98 84 1 2 0 3 1 2 0
t 0 0 16 3 0 4 1 0 1 0 2 10 0
|




